Day1- Introduction - data manipulation - graphical plotting
Walid Gharib - UniBe
Some words
‘R’ is a complete, flexible and open source system for statistical analysis which has become a core tool of choice for a wide range of researchers spanning over multiple disciplines. The aim of the present workshop is to give participants a first step hands-on practical session on the ‘R’ environment by introducing the basic principles and most used commands to help them explore and visualize their data.
What is R?
R is an open source complete and flexible software environment for statistical computing and graphics.
It includes:
- Facilities for data import, storage in memory and manipulation
- Functions for calculations on vectors and matrices
- Large set of data analysis tools
- Graphical tools
- As a programming language, a simple development environment, with a text editor
- R itself is written primarily in C and Fortran, and is an implementation of the statistical language S.
Advantages
- Availability and compatibility
- R is free and is documented (manuals, reference sheets, mailing lists, blogs, and a wiki)
- Can run on all major operating systems and code is often portable too
- R is free and is documented (manuals, reference sheets, mailing lists, blogs, and a wiki)
- State-of-the-art graphics capabilities
- R can produce quality graphical output in all of the standard formats (PDF, PS, EPS, JPEG, PNG, TIFF,…)
- Can import files from other statistical programs
- Such as Minitab, S, SAS, SPSS, or Stata (NOT from GraphPad Prism)
- Other file types such as tab-delimited text (tsv), comma-separated values (csv), and even Excel (xls) with proper packages
- Learn to program and do reproducible research
- It allows users to learn the basics of computer programming
- Writing scripts allows users to have a permanent, repeatable, annotated, cross-platform, shareable record of their analysis
- Automatic report generation that contain the code and the output of R scripts in various format (knitr and rmarkdown packages)
- It allows users to learn the basics of computer programming
- Speak the common language
- Knowing the basics of a tool will enhance the communication between collaborators and help in sharing information and data more easily
- R skills are in very high demand
- Knowing the basics of a tool will enhance the communication between collaborators and help in sharing information and data more easily
Drawbacks
- “Expert friendly”
- It is not very easy to learn and it takes time (steep learning curve)
- Needs prior formatting of the data
- (Too) large amount of resources
- Lots of documentation and manuals available, might be cryptic
- Sometimes easy to get lost
- Constantly evolving
- Can lead to backward-compatibility issues
- Memory intensive and slow at times
R-studio environment
- Launch it and let’s have a look
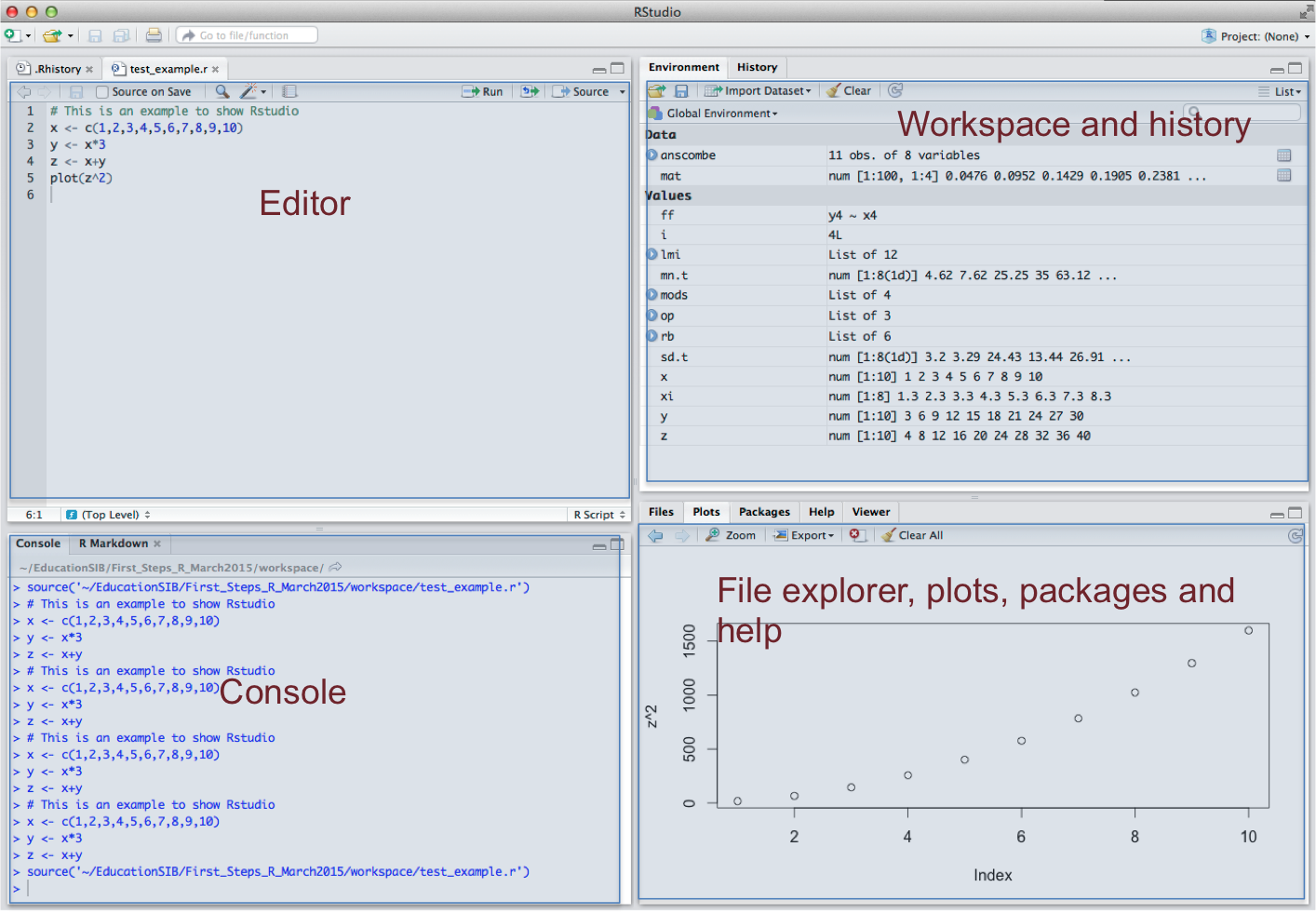
r-studio-env
Some environmental Functions
help()or?—> To ask for help !!!getwd()—> to print out the directory that is considered as working directory
setwd("path_to/the_directory/I_would_like_to/work_in")—> To set the directory that we would like it to be used as working directorysave.history()andload.history()—> The commands History so everything typed as commands will be remembered
save.image()andload.image()—> The environment will be saved not only the commands but also their output and all data loaded (size could become significantly big)
- Create an R script: from the upper menu select
File-->New File-->R Script, you can name it and choose the location Note To set your working directory without typing code, you can select from the upper menuSession-->Set Working Directory-->Choose Directory... - To execute a command from your R script, select the line or lines you want to execute and use the bottom
 located on the upper right corner of your script
located on the upper right corner of your script
- To comment a script with explanatory sentences you don’t want to execute you simply need to add the
#just before the sentence
Let’s warm up, ready!!!
On your own operating systems:
- Create a directory on your computers and name it
R-course
- Download the course materials provided on site or by email and put it in
R-coursedirectory you have just created
In R-studio:
- Create an R script and name it, save it in
R-coursedirectory
- Set your working directory to
R-courseby command line or by using the R-studio menu
setwd("/The/location/of/my/R-course.R")- Check what is your current working directory by typing the command in the R-script you have just created (From now on you will be typing commands in this area)
getwd()- Comment your script (at the top to start with)
# This a comment on top of my command
getwd()
getwd() # I can also comment my code at the level of my commandGeneralities
Calculator
4 + 2
4 - 2
4 * 2
4 / 2
4 ^ 2
sqrt(4)Assigning variables
a <-1
b <- "I am learning R - it looks cool !!"
bQ0- Assign the following “Hello everyone!” to a variable and call the variable on the next line
He<-"Hello everyone"
HeFunctions
v<-c(2,4,5) # the vector function c()
# We can also use functions over functions
mean(v)Printing output
print("Hello World")Data Types
Use the function class() to determine the data type of the following (N.B. assign every number to a variable before calling class() function):
- 7.5
a<-7.5
class(a)- 7
b<-7
class(b)- “whatever”
c<-"whatever"
class(c)- b+2i
d<-b+2i
class(d)- b>a
e<-b>a
class(e)- There are several logical operators
&,|,!among others.
f<-TRUE
g<-FALSEQ1- Run the two lines above and Try those operators on f and g and comment
f<-TRUE
g<-FALSE
f&g
f|g
!f
!gVectors
Integers c(1,3,5)
Booleans c(FALSE, TRUE, FALSE, TRUE)
String c("I","am","in","a","good","mood")
Q2- We could you use functions over vectors, for example: try the function
length()over the string vector above
length(c("I","am","in","a","good","mood"))Look at the following code:
functions as.factor(), levels() and nlevels()
v0<-c("I","am","in","a","good","mood","I","am","in","a","good","mood")
levels(as.factor(v0))
nlevels(as.factor(v0))Integers v1<-c(1,3,5) String v2<-c("I","am","in","a","good","mood")
Q3- Use the function
c()to merge vector v1 and v2
What do you notice?
v1<-c(1,3,5)
v2<-c("I","am","in","a","good","mood")
c(v1,v2)Q4- Create 2 integer vectors v3 and v4 of the same length and try some arithmetic on:
- calculate
mean(),median(),sum()
v3<- c(2,5,7,2,4)
v4<- c(23,66,224,89,65)
v3+v4
v3*v4
6*v3
v4/v3
mean(v4)
median(v4)
sum(v4)
#we could also do
mean(c(v3*v4))To Access a vector you could call the value position between square brackets e.g. vector[3] to call the 3rd value of vector
Q5- Access one value of the integer vectors v3,v4 and try some arithmetic functions between values from both vectors
- how do you proceed to retrieve the 2nd and the 4th position elements in your vector? or from the 2nd element till the last?
v3[2]+v4[5]
v4[c(2,4)]
v4[2:length(v4)]We could also add names to each value of the vector, e.g. if we have the height of 3 individuals of our family, we use the function names(), this function is also applicable to data.frame i.e. tables to output the column names.
height_family<-c(180,165,170)
names(height_family)<-c("Dad","Mom","Sis")
height_familylists
A list is very similar to vector while the only difference is that it can store multiple data types using the function list() for example :
x<-list("a",1,TRUE,1.5,1.5)Now let’s say we have multiple vector types with different length:
v5<-c("I"", "am", "gaining", "more", "knowledge", "in", "R")
v6<-c(1,2,3,4,5,6)
v7<-c(TRUE, FALSE, FALSE, TRUE)Q6- Load the vectors above by copy/pasting/executing them,
a-create a list l1 and store v5,v6,v7 in that list
v5<-c("I", "am", "gaining", "more", "knowledge", "in", "R")
v6<-c(1,2,3,4,5,6)
v7<-c(TRUE, FALSE, FALSE, TRUE)
l1<-list(v5,v6,v7)
l1b- Access and populate a list
To access a list values , we have to specify the index i.e. location of those values. for example to access the 1.5 value of our example list x all we need is to specify it’s location i.e. x[[c(4,5)]]
- Access The vector
v6located in the second position of the listl1 - Access the second element of the vector
v6from the l1 list
l1[[2]]
l1[[2]][2]Matrix
We can create a matrix using the function matrix() in which you create a vector of elements, the arguments are given nrow= (number of rows) and ncol= (number of columns) sand byrow=TRUE or FALSE (filling by row or by column)
for e.g.
v8<-c(2,5,9,1,3,4,8,7,0,12,14,16)
myFirstMatrix<-matrix(v8, nrow=3, ncol=4, byrow=TRUE)
myFirstMatrixQ7- If you noticed how the columns and the rows are represented (named) i.e. [,1], [,2] … for columns and [1,], [2,] for rows.
Using the indexes for rows and columns,how would you retrieve:
- the 3rd column from myFirstMatrix?
- the 1st row?
- the number 8 from the matrix elements?
- the number 16?
- the 1st and the 3rd column? note you should use the vector function
c()
v8<-c(2,5,9,1,3,4,8,7,0,12,14,16)
myFirstMatrix<-matrix(v8, nrow=3, ncol=4, byrow=TRUE)
myFirstMatrix[,3] # 3rd col
myFirstMatrix[1,] # 1st row
myFirstMatrix[2,3] # element 8
myFirstMatrix[3,4] # element 16
myFirstMatrix[,c(1,3)] # 1st and the 3rd colQ8- Try using the functions
mean(),median(),sum()on a row or a column ofmyFirstMatrixand finally trysummary()on myFirstMatrix
mean(myFirstMatrix[,3])
median(myFirstMatrix[1,])
sum(myFirstMatrix[2,] )
summary(myFirstMatrix)NOTE you can rename the rows and columns by using the function rownames() and colnames() for example
rownames(myFirstMatrix) <- c("firstR","secondR","thirdR")Q9- Rename the columns of myFirstMatrix
colnames(myFirstMatrix) <- c("firstC","secondC","thirdC","fourthC")Note You can add a column or a row to a matrix by using the functions rbind() and cbind() which takes as arguments the matrix and a vector or another matrix having the same length
for example:
newCol<-c(3,6,9)
cbind(myFirstMatrix,newCol)Q10- Add a new row to the matrix
newRow<-c(12,14,16,18)
rbind(myFirstMatrix, newRow)Data Frames - Diving into a real dataset
Data Frames are quantitative/qualitative data tables that are generally imported or loaded from various sources of information in the form of commas/tabulated/spaced/… separated fields.
About the dataset
Here we will dive into a real life example provided by the NHS National Health Service - England downloaded and adapted from Kaggle about Tobacco Use and Mortality in England between 2004-2015
All the needed information about the dataset can be found here
Download the dataset using this link
Installing external Libraries
Installing libraries in R is relatively simple, you just need to use install.packages() function and load in the package you wish to install e.g.install.packages('readr') or just doing it graphically by clicking on the packages option located on the right hand side in R-studio
readris a simple library that take care of importing csv, tsv, excel formats into R and load them as dataframes- For plotting an external famous library is required for the rest of the workshop simply because once you understand how it works it will make your life easier in “R” plotting compared to traditional way.
The library is called ggplot2 install it using packages in R studio or by using the command install.packages()
Data import
Import data using read_csv() function from the readr library
- To load an installed library into
Rall you need to use the functionlibrary()and load the library name as argument e.g.library(readr) - to import a csv (commas separated values) file pass the file name between quotes
""to the functionread_csv()
Note you can load a table using the interface Environment (upper right side of rstudio) panel by selecting  button
button
Admissions for smoking linked diseases
Q1-
a- Load the library ‘readr’ and import admissions.csv file (don’t forget to specify the location/path of the file admissions.csv, also put it between quotes "") load it into a variable which you will call admissions
library(readr)
admissions <- read_csv("~/Documents/RPPP/courses/teaching/Intro-to-R-decanatUnibe-2017/data/tobacco-use/admissions.csv")b- Use the function class()on the table admissions
class(admissions)
admissions<-as.data.frame(admissions)
class(admissions)c- Try the function summary() on admissions, comment
summary(admissions)let’s take some time too look at the table load it by typing View(admissions) in your console
Data selection rows and cols, reminder with matrix
Q2- Create a dataframe having only the first 3 columns of
admissionsI called mine admissions123
admissions123 <- admissions[,c(1,2,3)]as.date() example:
admissions$Year <- as.Date(admissions$Year, format='%Y/%d')
admissions$Year <- format(admissions$Year,'%Y') # extract only the year
head(admissions)data shaping
Almost all the dataset need to be shaped out for proper downstream analysis, why?
- empty fields
- format cells (numeric, dates etc)
- subsetting the data part we are interested in
We need to make some changes to the data.frame formats of some of it’s components. For example the year column should be formatted as Date using the function as.date() and the last column value should be formatted as numeric using the function as.numeric()
Q3-
as.numeric()is straight forward all you need is to pass the column you need to change it’s format as argument, try to do it yourself
admissions$Value<-as.numeric(admissions$Value)Q4- We need to replace the ‘NAs’ i.e. the cells that could be
MaleorFemalein the columnadmissions$Sexby another string and we are simply going to call it"MaleOrFemale", this is something you did not see before but I would like you to look at the code below, did you understand?
admissions$Sex[is.na(admissions$Sex)]<-"MaleOrFemale"Subsetting
Q5- Let’s say we are interested in looking at the number of hospital admissions of women and men as a total of all admissions regardless of the disease type between 2004 and 2015?
- If we look carefully at our admissions dataframe, for each year there is one row representing the number of admissions for diseases which can be caused by smoking, so we should subset these rows from the entire dataframe regardless of the sex. Try to do it by defining the column you and rows you need to answer the above question.
names(admissions)
adm_yr_allD<-admissions[,c(1,3,6,7)][admissions$`ICD10 Diagnosis`=="All diseases which can be caused by smoking",]
class(adm_yr_allD)Plotting using ggplot2
Here is a small intro to plotting using ggplot2 basic usage, let’s have a look:
Q6- Let’s open the code below and comment on each and every step - Don’t worry you will do it yourself on the next dataset
library(ggplot2)
admissions.Year<-adm_yr_allD$Year
admissions.Sex<-adm_yr_allD$Sex
admissions.Value<-adm_yr_allD$Value
women_men_admissions<-ggplot(adm_yr_allD ,aes(x=admissions.Year,y=admissions.Value,group=admissions.Sex,shape=admissions.Sex,colour=admissions.Sex)) +
geom_point() +
geom_smooth()
women_men_admissionsNote Try to replace geom_point() by geom_boxplot() or geom_violin() for different visualisation
Fatalities for smoking linked diseases
Q7- Now lets see if the same trend follows for the number of fatalities due to all diseases linked to cigarettes consumption.
- load in the file
fatalities.csvas done previously withadmissions.csv
fatalities <- read_csv("~/Documents/RPPP/courses/teaching/Intro-to-R-decanatUnibe-2017/data/tobacco-use/fatalities.csv") - Have a look at the loaded dataframe - It looks very similar to
admissionsso all manipulations prior to usage should be similar expect for thefatalities$Yearcolumn which looks a little different and needs to be converted toas.charater()becauseas.Date()do not operate on numeric()
fatalities$Year<-as.character(fatalities$Year)
fatalities$Year <- as.Date(fatalities$Year,format='%Y')
fatalities$Year <-format(fatalities$Year, format='%Y')Note We are showing those small conversions because in real life it is very rare to have a perfect dataframe ready to be used, data preparation is crucial before moving forward with any analysis
- Replace the NAs in
fatalities$Sexby'MaleOrFemale'similarly as done before
fatalities$Sex[is.na(fatalities$Sex)]<-"MaleOrFemale"- Set the Values in fatalities to numeric()
fatalities$Value<-as.numeric(fatalities$Value)- Subset the needed columns and keep only the cells that
'All deaths which can be caused by smoking'in'ICD10 Diagnosis'
names(fatalities)
fatalities_yr_allD<-fatalities[,c(1,3,6,7)][fatalities$`ICD10 Diagnosis`=="All deaths which can be caused by smoking",]- Using
ggplot()plot on the the years on the x-axis and the Values on Y axis
library(ggplot2)
fatalities.Year<-fatalities_yr_allD$Year
fatalities.Sex<-fatalities_yr_allD$Sex
fatalities.Value<-fatalities_yr_allD$Value
women_men_fatalities<-ggplot(fatalities_yr_allD ,aes(x=fatalities.Year,y=fatalities.Value,group=fatalities.Sex,shape=fatalities.Sex,colour=fatalities.Sex)) +
geom_point() +
geom_smooth()
length(fatalities.Sex)
women_men_fatalitiesNote if we want to plot only male and female we need to subset the data by only keeping male and female rows for example: fatalities_yr_allD_wo_MW<-subset(fatalities_yr_allD, fatalities.Sex %in% c("Female","Male"))
Note If you want to choose different colors you could add to the plot the function + scale_color_manual(values=c("yellow", "green", "orange"))